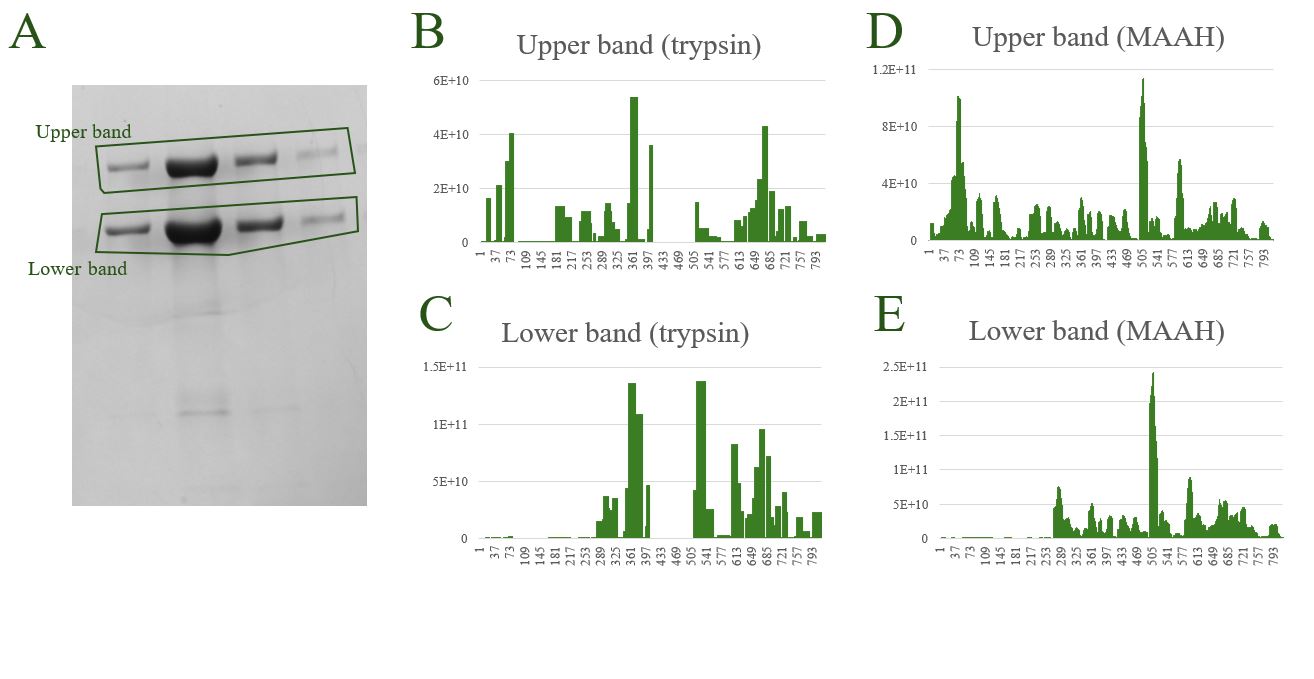
Isoform analysis
A common request is to characterize proteins – for example, protein bands that by SDS-PAGE are migrating faster or slower when compared to the expected protein band. For such questions high sequence coverage is often key. Using our most favored enzyme, trypsin that cleavages C-terminal to Lysine and Arginine we can often get an idea about what is going on. As an alternative we can use Microwave Assisted Acid Hydrolysis (MAAH) which provides a more ‘democratic’ cleavage of a protein. In the figure below are shown: A; two protein bands of interest, of which the faster migrating band is unexpected. Panel B and C shows plots of summed peptide signals as a function of the expected protein generated by trypsinization. In panels D and E are shown the corresponding analysis but with the samples subjected to MAAH.
While the trypsin experiment clearly shows missing N-terminal peptides for the faster migrating band (aka ‘lower band’) when comparing to the slower band, the MAAH experiment allows us to pinpoint the N-terminal of the faster migrating protein, that for this protein is Serine 271 flanked by a Methionine at the N-terminus. The MAAH experiment generated ~4,000 peptide ‘reads’ per sample while this number was ~200 for the trypsin digested samples. Samples generated by Nia Lyn@Marraffini.