Keeping telomerase in check
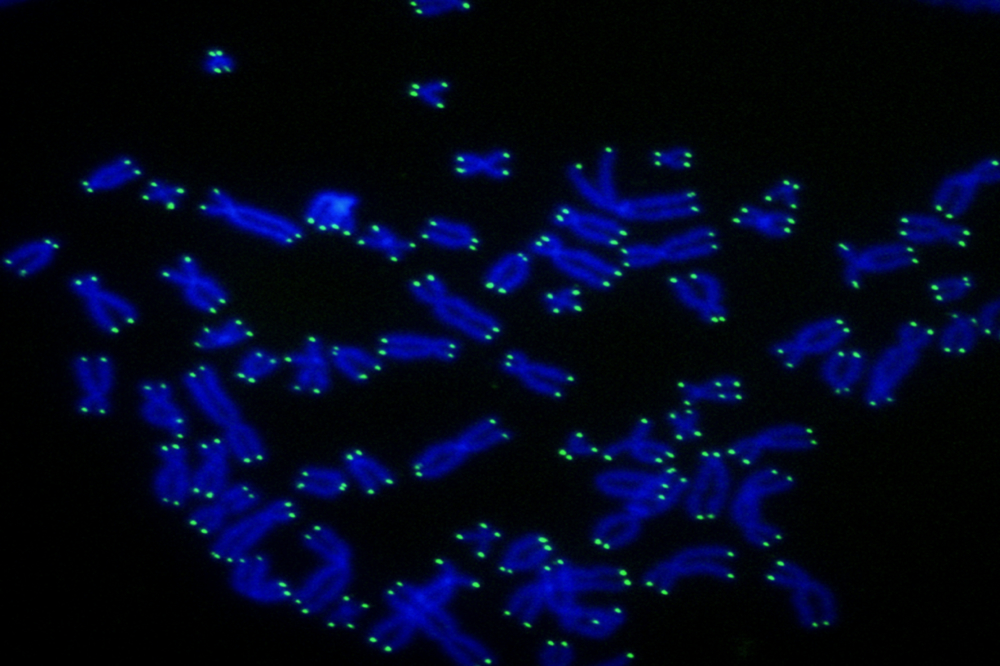
Human telomeres (green) at the ends of chromosomes (blue).
The natural ends of chromosomes appear alarmingly like broken DNA, much as a snapped spaghetti strand is difficult to distinguish from its intact counterparts. Yet every cell in our bodies must have a way of differentiating between the two because the best way to protect the healthy end of a chromosome also happens to be the worst way to repair damaged DNA.
Consider the enzyme telomerase, which is responsible for maintaining protective telomeres at the natural ends of chromosomes. Were telomerase to seal off a broken strand of DNA with a telomere, it would prevent further repair of that break and delete essential genes. Now, a new study in Science describes how cells avoid such mishaps. These findings show that telomerase can indeed run amok, adding telomeres to damaged DNA, and would do so were it not for the ATR kinase, a key enzyme that responds to DNA damage.
“Telomerase is a good thing because it maintains our telomeres, but it should only be acting at the natural ends of chromosomes. It is very bad if it acts at double-stranded DNA breaks because it can lead to the loss of all genes distal to the break,” says Titia de Lange, the Leon Hess professor at Rockefeller. “This detrimental aspect of telomerase is inhibited by the ATR kinase, which, among its many talents, also keeps telomerase away.”
The discovery may help optimize CRISPR techniques and could inform the study of cancer.
Enzyme vs enzyme
One of the earliest hints that telomerase could, absent proper controls, act on damaged DNA appeared in 1990, when a study in Nature reported that an individual suffering from α-thalassemia had a broken DNA end with telomeric DNA added to it. But whether telomerase was to blame for this rogue telomere, and how healthy cells prevented this from happening, remained unclear. Charles Kinzig, an MD/PhD student in the de Lange lab, scoured the literature for similar cases and set out to determine whether telomerase was the culprit.
Kinzig and colleagues first broke bits of human DNA with Cas9, the cutting component of the CRISPR gene-editing tool, and established that telomerase creates “neotelomeres” on broken DNA. Having established telomerase as driving the formation of neotelomeres, Kinzig then began interrogating various molecular pathways to determine what prevents telomerase from interfering with DNA repair under normal circumstances. He ultimately found that disrupting ATR kinase signaling increases neotelomere formation and demonstrated that when ATR is activated at DNA breaks, it prevents telomerase from ruining the repair.
“It’s a race between telomerase and ATR,” Kinzig says. “Telomerase needs the DNA end to be chewed in to form its single-stranded substrate. But at the same time, the single-stranded DNA is what activates ATR.”
From CRISPR to cancer
The findings have immediate implications for researchers and clinicians involved in CRISPR gene editing. Kinzig and colleagues found that telomerase can add telomeric DNA to the DNA ends made during CRISPR editing. This could potentially lead to insertion of telomeric DNA or formation of a telomere at the site where CRISPR editing was intended. “The CRISPR field now is aware of this and can take steps to prevent this unwanted outcome,” de Lange says.
In the long term, the lab plans to focus on how the findings relate to cancer. Telomerase is activated in most human cancers, and it is thought that this helps cancers maintain their telomeres, effectively becoming immortal. Kinzig and de Lange speculate that neotelomere formation may allow cancers to tolerate processes that generate broken chromosomes, such as deficiencies in BRCA1. “We are now testing whether neotelomere formation indeed helps cells deal with the genome instability that plagues cancer cells,” de Lange says. “We’ll see. Much remains to be learned.”
Kinzig, in the meantime, is finishing his medical training and preparing for the next step. “His thesis research was a tour de force,” says de Lange, “in part because he ventured into an area my lab had never worked on.”