New method gives scientists a better look at how HIV infects and takes over its host cells

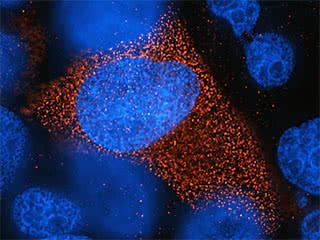
Cell to cell: When HIV infects a cell, it programs the cell to express the viral protein Env, which the virus uses to spread to neighboring cells. Above, Env (red) produced by one infected cell has recruited other, uninfected cells, causing them to fuse, and their nuclei (blue) to cluster.
Viruses attack cells and commandeer their machinery in a complex and carefully orchestrated invasion. Scientists have long probed this process for insights into biology and disease, but essential details still remain out of reach.
A new approach, developed by a team of researchers led by The Rockefeller University and The Aaron Diamond AIDS Research Center (ADARC), offers an unprecedented view of how a virus infects and appropriates a host cell, step by step. In research published May 23 in Nature Microbiology, they applied their method to HIV, a virus whose genome is less than 100,000 times the size of ours.
“HIV is truly an expert at living large on a small budget,” says first author Yang Luo, a postdoc at ADARC and a former graduate student at Rockefeller University. “We asked the question, how does such a compact virus manipulate the host cell to gain entry and replicate itself, all while escaping the immune system?”
Mapping HIV’s “interactome”
The study focused on two viral proteins known to bring about HIV’s infection of human white blood cells. The first one, called envelope or Env, sits on the surface of the virus and, by binding to receptors on the host cell, helps the membrane that encapsulates the virus fuse with the cell’s outer membrane. A second protein, Vif, destroys an enzyme that host cells produce to defend themselves against the virus.
In an effort to better understand how these two proteins function, the team wanted to map their interactome—meaning all the proteins with which they associate within a host cell. To accomplish this, the researchers needed to devise a way to isolate clusters of interacting proteins from cells during different stages of infection. Such experiments can be done by introducing a genetic sequence into the viral genome—a “tag” that acts like a piece of molecular Velcro, allowing one viral protein to be yanked out along with all the other proteins associated with it.
Molecular arms race: The HIV Vif protein coordinates the destruction of a host cell’s defenses. Above, 22 hours after the start of an infection, speckles of Vif (red) are visible in the cytoplasm of an infected cell.
It sounds simple, but making it work took a decade.
“Inserting a tag sequence into small viruses is a challenge to begin with,” says corresponding author Mark Muesing, a principal investigator at ADARC. “If you disrupt their nucleic acid and protein sequences, you can easily compromise the virus’s ability to replicate. And HIV represents a particular challenge because it can quickly revert back to its original sequence.”
“We developed a technique to find places in the HIV genome where we can insert stable tags without affecting the virus’s capacity to proliferate. In effect, this allowed us to expand cultures of the infected cells along with the tagged viral protein,” he adds.
The host’s contribution
Next, the researchers infected human cells with viruses carrying the tagged protein sequences, and were able to pull out and identify a large number of host proteins directly during the infectious process. This provided the first evidence that many previously underappreciated host proteins interact with the viral machinery during replication.
“Imagine you have a factory assembly line where only one component of, say, the stamping machine, actually touches the product,” says co-author Michael Rout, professor and head of Rockefeller’s Laboratory of Cellular and Structural Biology. “Other parts support and power the stamp. Likewise, within an infected cell, we can identify the components of a particular cellular machine, not just the piece that comes in contact with the viral protein.”
“Every host protein we pull out generates new questions,” adds co-first author Erica Jacobs, a research associate in Brian Chait’s lab. “Does it help the virus invade and co-opt the host to replicate itself? Or does it harm it? The answers will not only help us understand the virus, but also shed light on our cells’ ability to defend themselves.”
One important discovery has already emerged from the lists of proteins. Viruses, including HIV, often attack as so-called virions, which are individual packets of protein and genetic code. But they can also pass directly from an infected to an uninfected cell, a more effective mode of transmission. To do so, the virus appears to use host proteins to construct a platform, a close junctional surface, between cells.
From the list of proteins that interact with Env, the researchers have identified cellular proteins predicted to contribute to this platform between cells. Because this route of transmission protects the virus in a sequestered environment, away from host defenses, the new findings may aid in the development of future anti-HIV therapies.
A live infection, step by step
According to co-author Brian Chait, Camille and Henry Dreyfus Professor and head of the Laboratory of Mass Spectrometry and Gaseous Ion Chemistry, the new approach offers a rare glimpse into the process by which HIV invades and resurrects itself within a cell.
“Often, studies of this sort are done with viral proteins in the absence of a true viral infection,” says Chait. “However, because viral infections are exquisitely orchestrated events, you are likely to miss all kinds of important details if you study the action of these proteins out of their proper context.”
“Deciphering the intricacies of virus–host protein interactions in space and time during the progression of an infection is remarkably powerful,” says co-author Ileana Cristea, an associate professor of molecular biology at Princeton University. “The challenge is to discover which precise interactions are the critical ones.”
Todd Greco, a co-first author, and an associate research scholar and lecturer in molecular biology in Cristea’s lab, says that “even for host proteins within the same family, their relative stability within HIV-1 protein complexes can be very different. More broadly, by understanding these mechanisms we will better understand the coordinated responses of cells.”
This work was supported by the National Institutes of Health (grants: R01AI047054, R21AI065321, R01AI081615 and R21AI097233, P41GM103314, U54GM103511, P41GM109824, R01AI093278, R33AI084714, DP1DA026192, and R21AI102187).
 Nature Microbiology, online: May 23, 2016 Nature Microbiology, online: May 23, 2016HIV–host interactome revealed directly from infected cells Yang Luo, Erica Y. Jacobs, Todd M. Greco, Kevin D. Mohammed, Tommy Tong, Sarah Keegan, James M. Binley, Ileana M. Cristea, David Fenyö, Michael P. Rout, Brian T. Chait, and Mark A. Muesing |