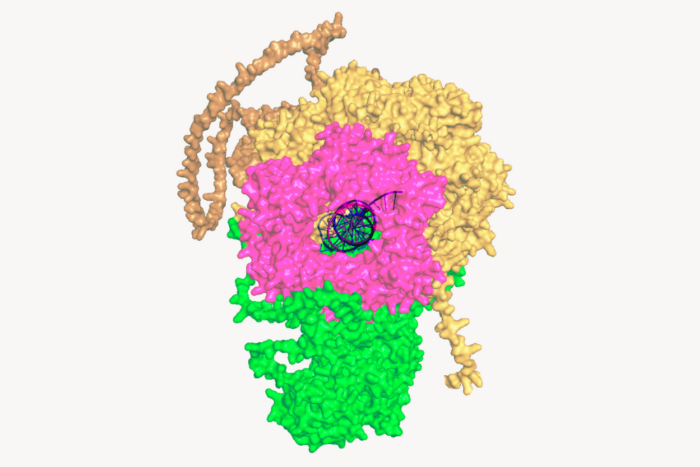
A new method reveals hidden rules of gene control
Key takeaways
- Scientists struggle to see direct control of gene expression in cells, because interfering with the system often obscures the very effects they hope to measure.
- A new method that rebuilds transcription outside of the cell can show scientists which genes are directly controlled by specific factors.
- By revealing the direct logic of gene regulation, the new method could reshape our understanding of biology, and help uncover new vulnerabilities in dangerous pathogens.
An artistic rendition of Mtb transcription. (credit: Andrew Tivon)
Inside every cell, thousands of molecular signals collide, overlap, and compensate, obscuring the true drivers of gene expression. Scientists have now developed a way to silence that cellular noise, revealing transcription drivers by reconstructing transcription outside of the cell.
This approach, described in a paper published in Molecular Cell, focuses squarely on how the enzyme that copies DNA into RNA operates to allow unique insight into how genes are switched on and how RNA synthesis begins and ends. Researchers used this method to reveal fundamental features of the transcription cycle—the process by which cells copy DNA into RNA to make proteins—in Mycobacterium tuberculosis (Mtb). The findings may help scientists better understand this pathogen, as well as the drugs designed to combat it.
“Gaining a deep understanding of how transcription works reveals central principles in biology that have huge significance for human health,” says Elizabeth Campbell, head of the Laboratory of Molecular Pathogenesis. “We can more precisely design therapeutics to target a process if we truly understand it.”
The search for direct effects
Bacteria like Mtb survive by precisely controlling which genes are turned on or off in response to changing conditions. At the center of this process is RNA polymerase (RNAP), which copies DNA into RNA, and a troupe of transcription factors that interact with RNAP to fine-tune gene activity. But these interactions are buried in dense, noisy networks within the cell—an environment that obscures the direct effects of each transcription factor.
To understand what a transcription factor specifically controls, scientists typically weaken or remove it and then measure which genes change their activity. In theory, those shifts should reveal the transcription factor’s direct targets. But in many living cells, including Mtb, disrupting an essential transcription factor triggers emergency compensation or cellular collapse, unleashing widespread ripple effects that drown out the transcription factor’s original signal. Meanwhile, standard genomic tools capture only fragments of direct effects. The ChIP-seq method reveals where a protein binds but not whether or how it alters gene activity; the RNA-seq method reveals which genes change after a disruption but not whether those changes are direct or indirect.
“We cannot identify direct targets this way. We’ve tried and tried, and many others have tried,” Campbell says. “If gene expression is a pathway then, when we use these methods, we’re just seeing the endpoint. We’re never seeing what’s happening along the pathway.”
A new method was needed, in part to enable the study of the many organisms that cannot be cultured in the lab. “In order to really uncover general principles, one must be able to study samples across different phyla of bacteria, including those that we cannot culture,” Campbell says. That urgency was magnified in Mtb. Without defining the direct effects of its transcription factors, researchers are limited in fully understanding how to fight back against that pathogen.
The Campbell lab wondered whether transcription could be rebuilt and studied outside the cell. A controlled, cell-free system might reveal the direct effects of a transcription factor without interference. Such a method could help uncover the rules of transcription across diverse species, from Mtb to organisms that cannot be cultured at all.
Cell-free genomics
The effort to build such a method was spearheaded by Ruby Froom, then a graduate student in the Campbell lab. “I had the idea to see if we could reconstitute the system in a test tube and identify direct targets outside the cell,” Campbell recalls. “But genomics isn’t my expertise. It took Ruby’s creativity, rigor, and persistence to transform that idea into a working platform.”
To reconstruct the entire process in a controlled, cell-free genomic system, Froom used purified components from Mtb. The team extracted and fragmented DNA, then combined it with RNAP, its primary sigma factor, transcription factors, and specific regulators including CRP, WhiB1, NusA, and NusG. By running parallel reactions with and without individual factors, they isolated each protein’s direct impact on RNA synthesis and then designed sequencing approaches to identify the exact genomic positions where transcription starts and stops, allowing them to map the precise activity of RNAP. Using custom computational analyses, the researchers then quantified how each factor changed gene activity and identified the DNA patterns that drove those changes. They cross-checked the results in living cells and validated key predictions with precise, single-gene experiments.
They found that the system revealed fundamental rules of how Mtb controls its genes. With their new method, the Campbell lab showed that the bacterium’s transcription machinery relies on classic DNA start signals that had appeared weak or absent in living cells, suggesting those signals have until now been masked. The team was also able to map the complete set of genes directly controlled by a well-known regulator called CRP, revealing dozens that it governs independently, without help from other cellular factors. In some cases, the method even separated true cause from collateral damage, showing that what may seem like global regulators in living cells are often far more precise. The team found that the transcription factor WhiB1, for instance, directly controls only a small set of critical genes, even though disrupting it in living cells unleashes widespread chaos.
This cell-free approach also helped the team resolve a long-standing debate about how transcription ends, showing that sequence-driven termination operates across the Mtb genome and clarifying the distinct roles of NusA and NusG. “NusG is the only transcription factor conserved across all domains of life, from bacteria to humans,” Campbell says. “These findings position Mtb as a powerful system for uncovering universal principles of gene regulation.”
While powerful, Campbell stresses that the new method is meant to augment, rather than replace, existing techniques. “Our approach complements current genomics methodologies, to address the direct effects of transcriptional factors,” Campbell says.
The implications of the new method extend beyond basic biology. RNA polymerase is the target of rifampicin, a frontline tuberculosis drug, and this new method may help researchers better understand precisely how it operates—offering new insight as resistance rises.
Beyond tuberculosis, the study implicitly questions the long-standing reliance on classic model organisms such as E. coli to define the basic rules of gene regulation. By examining transcription directly in a different organism, the work suggests that important aspects of gene control can remain hidden when scientists rely on only one experimental framework.
“With this method, we are not just trying to see how Mtb compares to standard models. We are establishing new principles of gene expression, and uncovering ones that haven’t been addressed in other organisms,” Campbell says. “There is no one ‘model’ anymore. Just as there is no model human or model culture, bacteria are all different. We should study it all.”