Scientists capture the moving parts of the portal to the cell’s nucleus
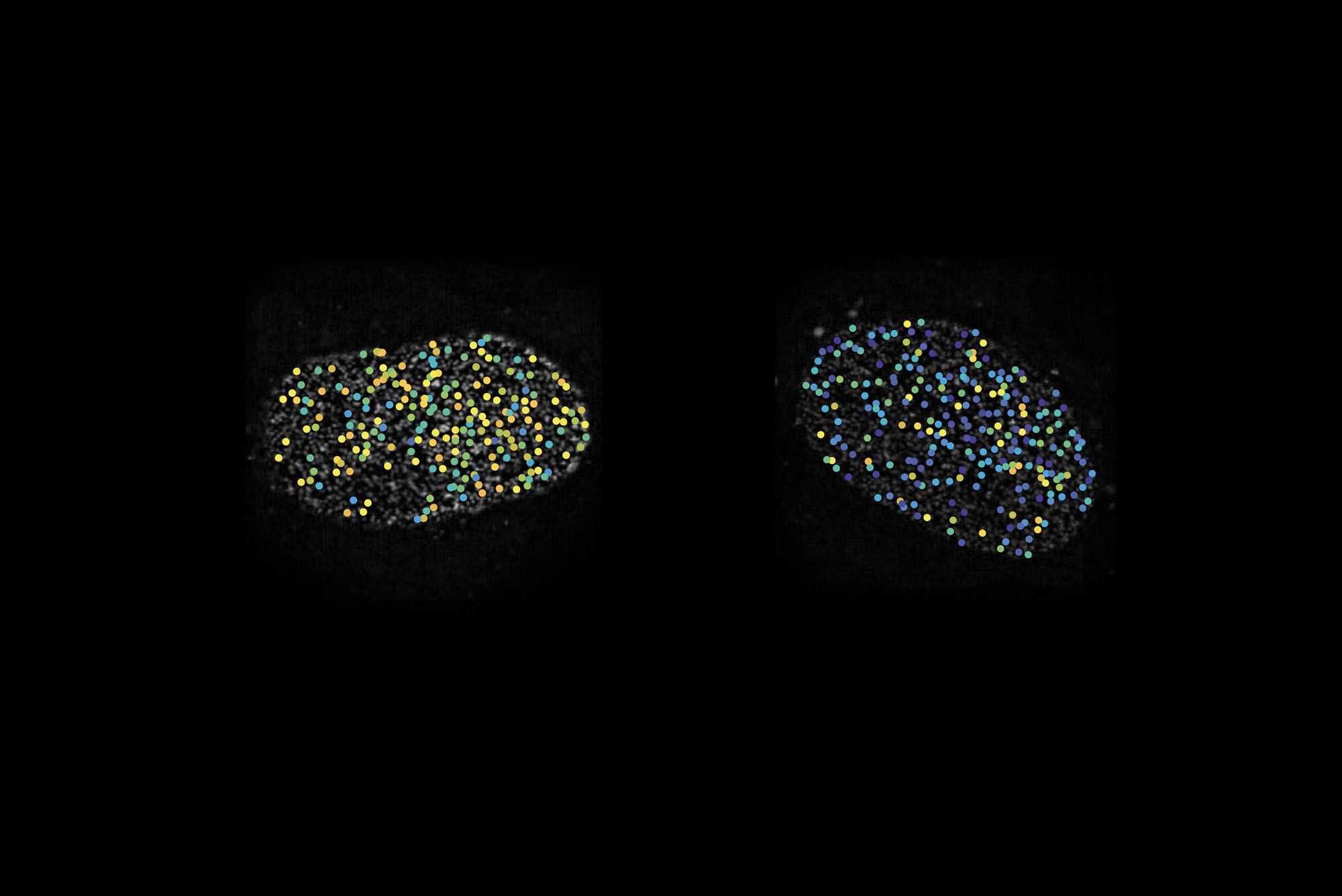
Two cell nuclei whose nuclear pores appear as dots. The colors represent different orientations of a key protein in the pore.
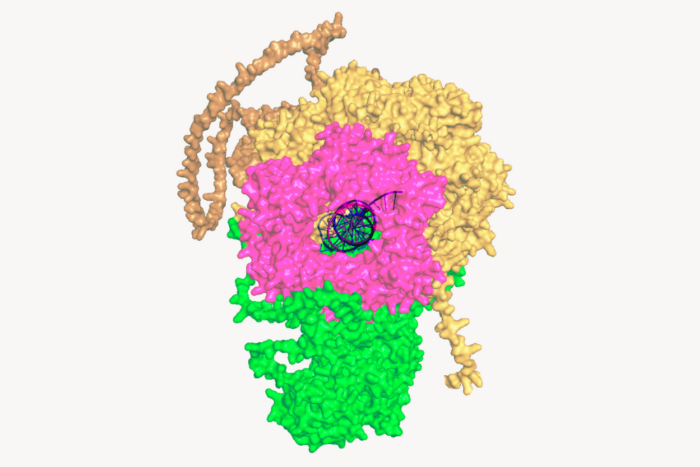
The nucleus of a cell is well-shielded by a double membrane to protect its most sensitive possession—its DNA. Anything that enters or exits must pass through the nuclear pores, cylindrical structures made of hundreds of proteins. Scientists have made great advances in figuring out the architecture of the pore by reconstructing each of its parts in the lab. But they have struggled to understand how those parts work together—whether the pore is a passive drain or a dynamic, flexible structure.
To find out, researchers at Sanford M. Simon‘s lab at Rockefeller developed a way to track the movements of proteins in the nuclear pore when it’s doing its job inside a living cell. Their findings, published in eLife, suggest the proteins forming the pore’s inner ring constantly change their orientation as they let cargo through, changing the whole conformation of the channel.
“It suggests it may be working like a finger trap toy,” Simon says.
For the study, the researchers embedded fluorescent chemicals in the nuclear pore proteins. These markers brighten up only when light is shone onto them at an angle parallel to their direction, so by monitoring the light they give off, the researchers can infer the precise orientation of a protein, or even subdomains of a protein.
The team then manipulated the amount of cargo transport through the pore and monitored the motion of the proteins. While most compartments of the nuclear pore were static, the inner ring proteins flip-flopped rapidly between two orientations. “If we shut down transport, they would stay in one orientation. If we started it up, they would switch,” Simon says.
The technique, called fluorescence polarization microscopy, can be used to study many other molecular structures with moving parts. “A structure can behave quite differently when it’s taken out of its native context,” says Joan Pulupa, a former graduate student at Simon’s lab, now a postdoc at Columbia University. “With this approach we can study structures not just in vitro, but also inside living cells while they are active and doing work.”