Cell cycle control involves coordinated production and destruction of proteins that activate cyclical events required for precise cell duplication. Cross uses a variety of approaches to investigate cell cycle control at the molecular level.
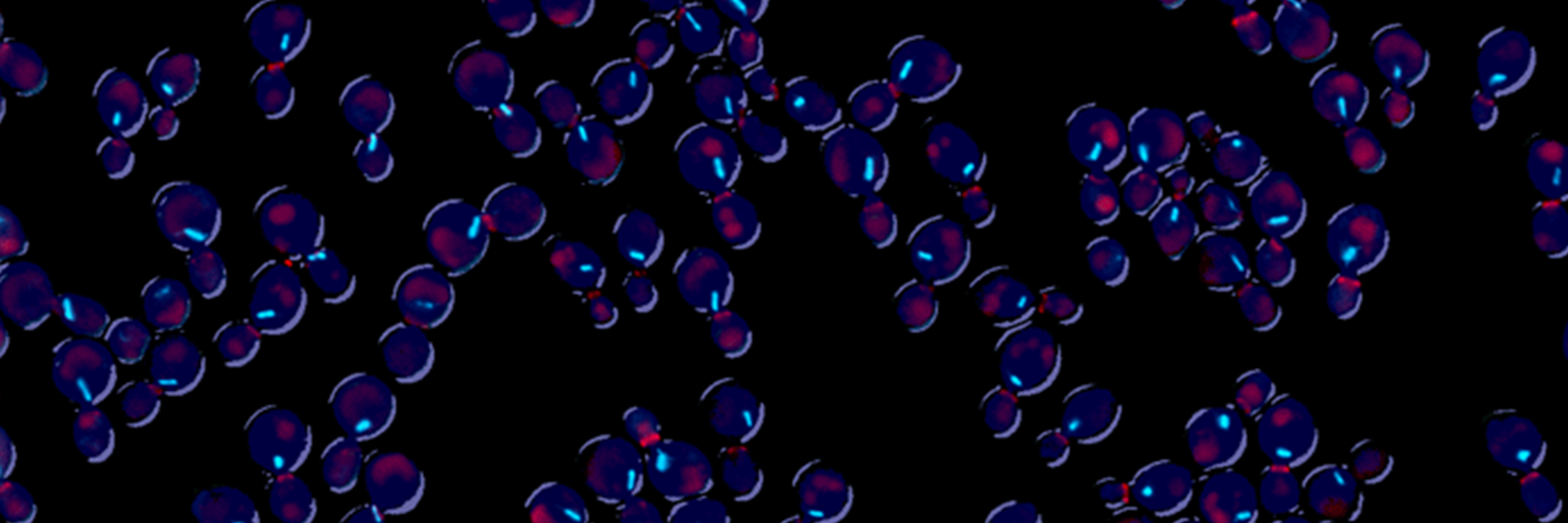
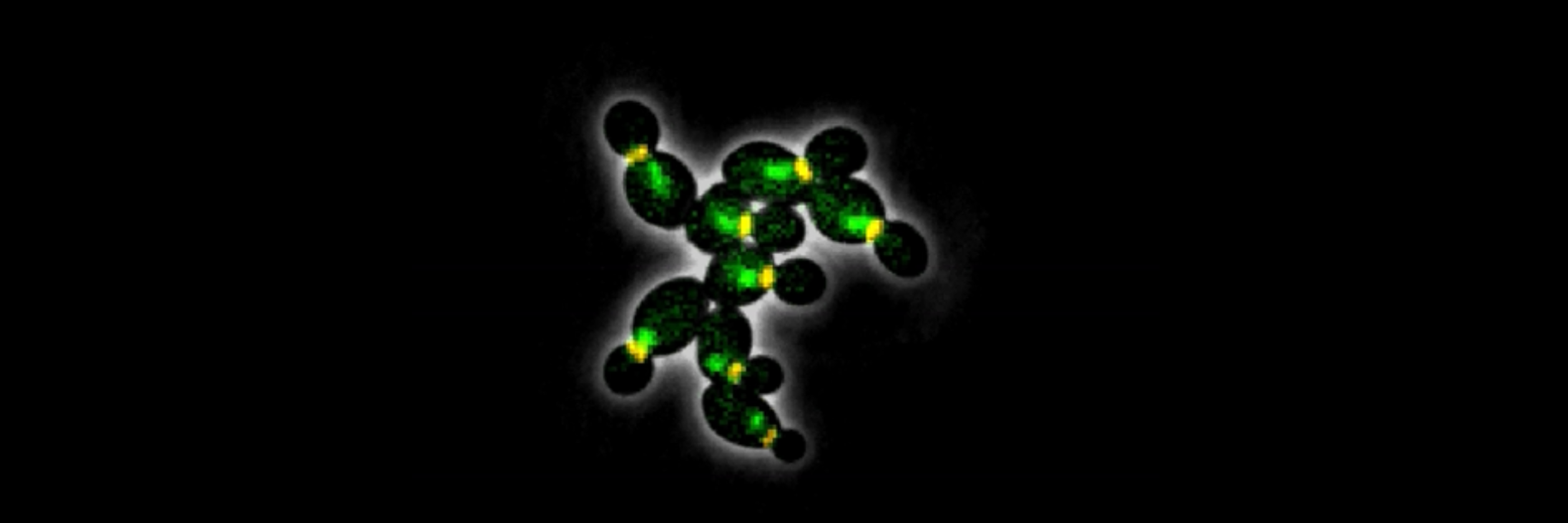
Using budding yeast as a model system, Cross uses both genetic and biochemical approaches to investigate the molecular basis of cell cycle control. He seeks to understand how critical regulatory proteins called cyclins control cell cycle progression, both through their timely degradation and through their ability to be highly selective of the molecules with which they interact.
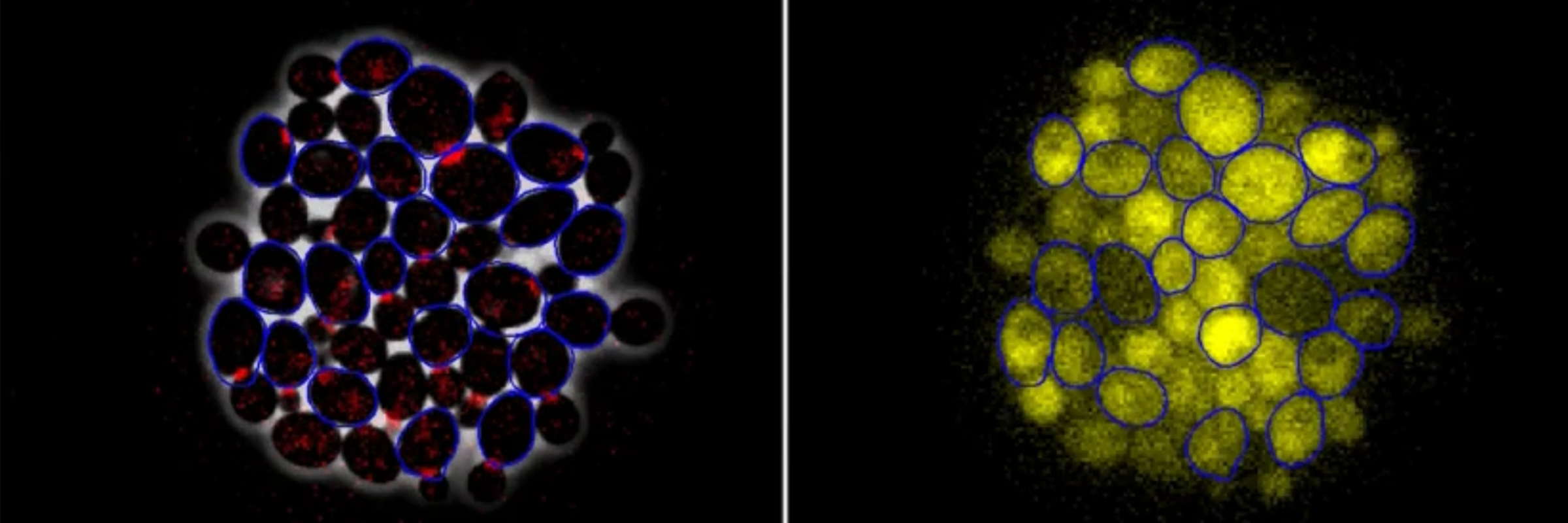
The laboratory is interested in systematic approaches to cell cycle control, including mathematical modeling. Researchers in the Cross lab are developing mathematical models that represent control of the cell cycle and are creating single-cell imaging methods for regulation of gene expression and protein localization through the cell cycle.
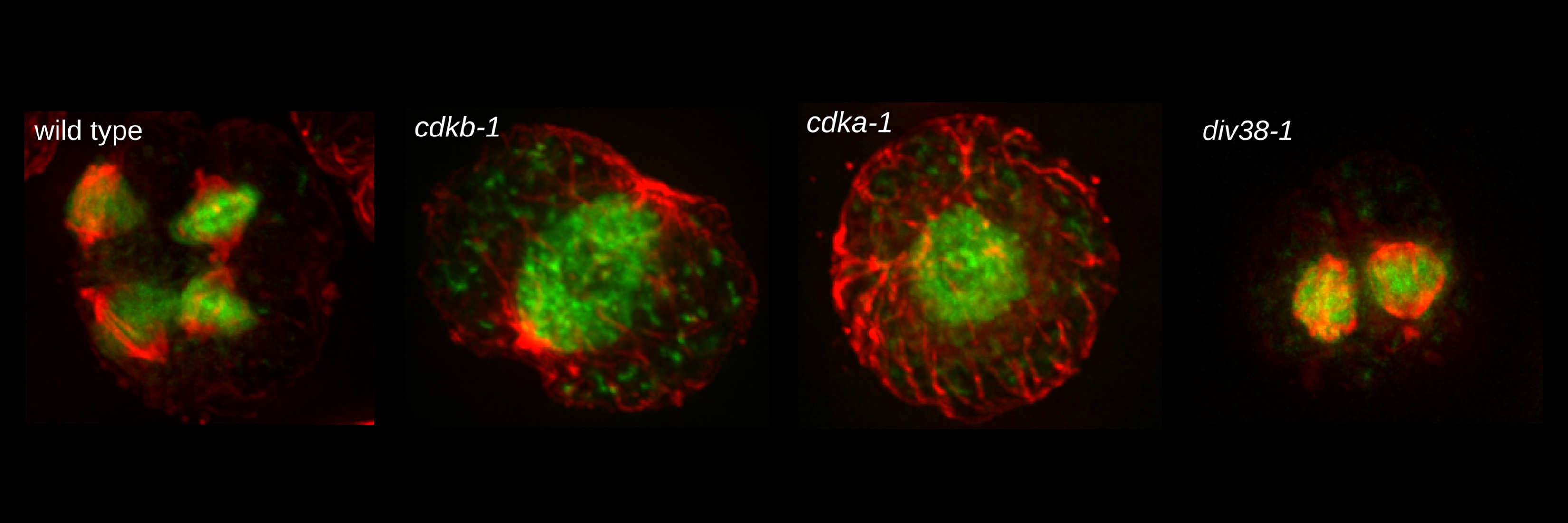
A second project concerns cell cycle control in the green alga Chlamydomonas, which is a good microbial genetic model for the plant superkingdom. Plant genetics is challenging because of long generation times, diploid genetics, and ancient polyploidizations that result in many genes being present in multiple functional copies, masking loss-of-function phenotypes. Chlamydomonas, with essentially a full plant genome with respect to core cell biology including cell cycle regulation, has almost all of its genes in single copy, is haploid, and is amenable to classic microbial genetics, as well as modern molecular methods. The lab is creating a systematic collection of mutations in all genes involved in Chlamydomonas cell cycle control, and is using these mutants and other tools for focused studies on similarities and differences in eukaryotic cell cycle control across kingdoms.
