Scientists have gotten good at blocking enzymes to treat disease. Now can they speed them up?

Tarun Kapoor (Credit: Lori Chertoff)
Enzymes are the molecular machines that power life; they build and break down molecules, copy DNA, digest food, and drive virtually every chemical reaction in our cells. For decades, scientists have designed drugs to slow down or block enzymes, stopping infections or the growth of cancer by jamming these tiny machines. But what if tackling some diseases requires the opposite approach?
Speeding enzymes up, it turns out, is much harder than stopping them. Tarun Kapoor is the Pels Family Professor in Rockefeller’s Selma and Lawrence Ruben Laboratory of Chemistry and Cell Biology. Recently, he has shifted the focus of this lab to tackle the tricky question of how to make enzymes work faster. Already, his lab has developed a chemical compound to speed up an enzyme that works too slowly in people with a rare form of neurodegeration. The same approach could open new treatment possibilities for many other diseases where other enzymes have lost function, including some cancers and neurodegenerative disorders such as Alzheimer’s.
We spoke with Kapoor about why speeding up an enzyme is so difficult, and how his team is cracking this problem.
Your lab spent about two decades studying cell division. What led you to shift focus?
This newer work really builds on my training. I did my PhD in chemistry and structural biology, focusing on designing small molecule inhibitors—compounds that block protein function. Then in my postdoc, I worked in a cell biology lab where we specifically developed small molecule inhibitors of cell division. Drugs targeting this process have been a mainstay of cancer chemotherapy for many years.
My lab has been very productive in that area for twenty years, and many of my former trainees—24 are now professors—continue to work on cell division. But I started asking what else we could do with the tools and approaches we’ve developed over the years. And I thought, we’ve gotten really good at stopping enzymes. But could we make them go faster?
That question led us to focus on ATPase enzymes—molecular machines that use chemical energy (ATP) to do mechanical work, like unfolding proteins or transporting them around the cell. These enzymes are involved in so many processes beyond cell division: immune cell function, managing the life cycle of proteins, even viral replication. And it turns out that for many diseases, such as neurodegeneration, the problem isn’t that these enzymes are working too well, but that they’re not working well enough.
Why is speeding up an enzyme so much harder than inhibiting one?
Think of it like a bicycle. If I want to completely stop a bicycle, I can come up with lots of ways to do that. I can jam the wheel, cut the chain, whatever it takes to break it down or block its movement. But now imagine the converse: I want to make a bicycle go faster. It’s more complicated now. How much faster does it need to go? Will it tip over or veer off course if it goes too fast? Where’s the gearbox that needs to speed up? There’s more nuance required to achieve the result without breaking the whole system.
That’s the situation with enzymes. We’ve been very successful at shutting them off because we can use blunt tools to break an enzyme, but it takes a much deeper understanding of these molecules—which comes from basic science research—to speed them up.
You’ve been working on a protein called VCP. Tell us about this enzyme and why it’s an important target.
VCP is essential for controlling the life cycle of proteins. Proteins are constantly being made and destroyed in our cells, and VCP plays a critical role in managing that process. It’s like sorting damaged goods: do you send them to be recycled or to the trash? VCP helps make those decisions and prepares proteins for whichever fate they’re headed toward.
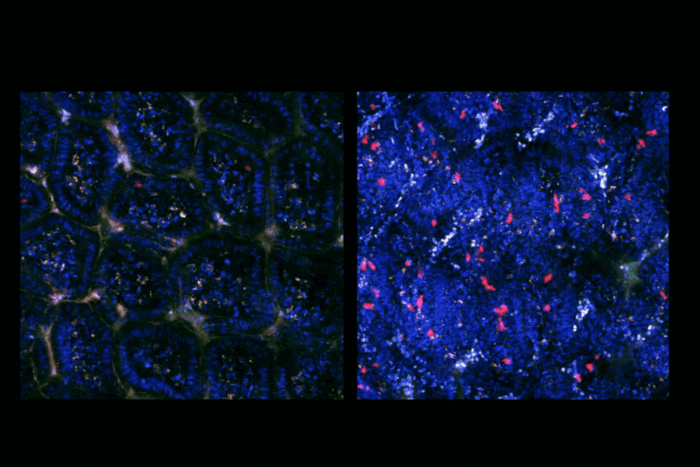
Patients with VCP mutations can develop a rare, debilitating condition that makes proteins accumulate when they should be discarded by the cell. Over time, this leads to neurodegeneration and dementia. The hypothesis is that in these patients, the enzyme is still present and active, but it’s working more slowly than it should.
How did you go about trying to make VCP work faster?
We didn’t know how to control the speed of VCP, so we started with an unbiased approach. We screened about 30,000 drug-like compounds in test tubes using a purified protein, testing whether any of them made VCP work faster. We found a few hits, then used chemistry to modify and improve them, which was a good sign that we were onto something.
The real breakthrough came when we used cryo-electron microscopy to see exactly where and how these compounds bind to the enzyme. We discovered a kind of gearbox in the enzyme. The compound essentially shifts the enzyme into a higher gear by binding to that area of the enzyme.
Now that we know the importance of that gearbox, we can work on further improving our activating compound very quickly by determining how well new variations of the compound bind to that gearbox and how that changes VCP’s activity.
Do you think what you’re learning will apply to other enzymes?
The specific gearbox we found in VCP probably won’t be identical in other enzymes, which is actually a good thing for drug development; you want drugs to be able to target one enzyme without speeding up all the others in the body. But the basic approach and the conceptual framework should be broadly applicable.
There are many diseases where proteins aren’t working well enough—what we call loss of function. You might lose one copy of a gene and only make half as much of a protein, or you might have a mutation that makes the protein work more slowly. For instance, there’s a drug currently in clinical trials that treats heart failure by improving the work of an enzyme that’s key to the heart’s ability to contract.
If we solve one problem well, if we could help treat even one disease, that would be valuable. Then we can apply those lessons to the next enzyme, and the next. Over time, patterns should emerge.
Can AI help find these patterns and design new enzyme activators?
Right now, AlphaFold and similar AI tools are incredibly powerful for predicting what proteins look like. But they can’t really help us here because enzymes are constantly moving and changing shape as they do their work. It’s like the difference between having a photograph of a bicycle versus understanding how it actually functions.
For understanding enzyme mechanisms and finding ways to modulate their speed, we need to capture the enzyme in action, in multiple different states, and piece together how it’s moving through its cycle. That requires experimental approaches—biochemistry to measure how fast the enzyme works, structural biology to see where compounds bind, and proteomics to track what happens in cells. That’s where my lab is focused.