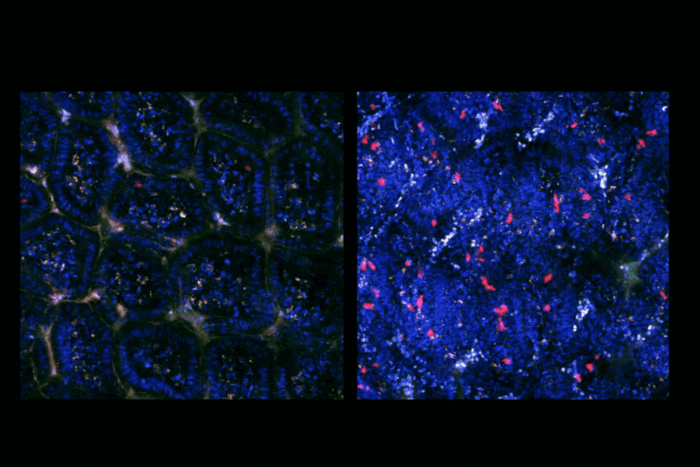
New faculty member unlocks vast chemical spaces for drug discovery
 Somewhere in every biology question, there’s a lock-and-key problem. To understand how cells communicate and function, or to develop new drugs, scientists must identify molecules that bind to the cells’ receptors—an immense task. There are myriad receptors, and the keys that unlock each of them—their so-called ligands—might be any of billions of molecules out there.
Somewhere in every biology question, there’s a lock-and-key problem. To understand how cells communicate and function, or to develop new drugs, scientists must identify molecules that bind to the cells’ receptors—an immense task. There are myriad receptors, and the keys that unlock each of them—their so-called ligands—might be any of billions of molecules out there.
Jiankun (JK) Lyu, the newest addition to The Rockefeller University faculty, is a computational chemist at the forefront of conducting the search computationally. He develops algorithms that excel at finding the elusive molecules that bind to a given receptor and make it possible to expand the search to libraries of millions to billions of molecules.
“The chemical space is vast, and we have only explored a fraction of it,” says Lyu, who is currently a postdoc at University of California, San Francisco (UCSF). “My goal is to increase the size and diversity of the chemical space that we are able to screen.” Lyu will join Rockefeller January 1, 2023, as assistant professor and head of the Laboratory of Computational Molecular Discovery.
The ability of a molecule to dock itself into a protein’s deep cavities depends on its structure and chemical properties. Increasingly, researchers use computational tools and specialized software to analyze the properties of molecules and predict the ones with the highest chance of forming a stable complex with a protein target—a mark of the molecule’s potential as a future drug.
From the very first time he saw a drug-protein complex on a computer screen, Lyu was hooked. “I was astonished. It looked so elegant,” he says. During his graduate studies at the East China University of Science and Technology, and especially later in the lab of Brian Shoichet at UCSF, where he finished his Ph.D. and continued as a postdoc, Lyu worked on improving computational methods and algorithms for analyzing small molecules and their interactions with protein targets.
Besides making the algorithms faster and more reliable, Lyu was also interested in improving the process on another front: by increasing the size and diversity of the search space itself. One great advantage of computational methods is that they make it possible to screen beyond the usual suspects and find new or improved drugs by screening chemicals that may look too different to a protein’s known ligands, but in fact work in a similar way.
Lyu believed, and later showed in his studies, that enlarging the library of molecules available for virtual screening will increase the chances of a successful find. In some cases, a molecule that looks nothing like the protein’s existing ligands may turn out to be just as potent, or even outperform them.
To that end, he developed an innovative computational platform capable of screening billions of small molecules, known as ‘ultra-large library docking.’ The best hits identified on this platform are then purchased or custom-made by chemical synthesis companies, so actual binding ability of the physical molecules can be tested in the lab. To showcase this approach, Lyu used it to search hundreds of millions of molecules and successfully identified novel ligands for the dopamine and sigma receptors, both of which are targets for psychoactive drugs.
“JK’s findings illustrate the immense opportunities for rapid discovery of ligands using structure-based screens of diverse, ultra-large virtual libraries, and have led to an incredible expansion of the molecules and chemotypes available to the chemical biology and drug discovery communities,” says Richard P. Lifton, Rockefeller’s president. “His ingenuity and out-of-the-box thinking coupled with his leadership in computational biology make him a terrific addition to our faculty.”
At Rockefeller, Lyu is planning to continue developing faster and more powerful computational methods and pushing the limits of the space of molecules that can be explored. He also plans to expand the application of these methods to finding ligands for engineered or designed proteins, providing scientists with new layers of control over the proteins that are commonly used as chemogenic tools and biosensors to study vast areas of biology and disease.